The WES R package provides reproducible functions for collating and analyzing data from wastewater and environmental sampling studies. Wastewater and environmental sampling (WES) of infectious diseases involves collecting samples from various sources (such as sewage, water, air, soil, or surfaces) to monitor the presence of pathogens in the environment. Analysis of WES data often requires the calculation of real-time quantitative PCR (qPCR) variables, normalizing WES observations, and analyzing sampling site characteristics. To help reduce the complexity of these analyses we have implemented tools that assist with establishing standardized WES data formats, absolute and relative quantification of qPCR data, estimation of qPCR amplification efficiency, and collating open-source spatial data for sampling sites.

Features
Features of WES include:
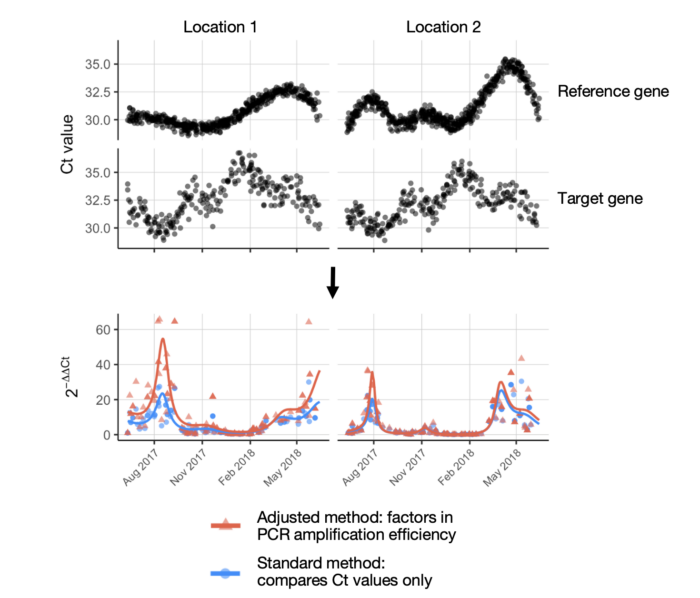
- qPCR data analysis: It provides functionality for calculating real-time quantitative PCR (qPCR) variables, including absolute and relative quantification, which are essential for analyzing pathogen presence in environmental samples.
- Standardized WES data formats: The package helps in establishing standardized data formats for wastewater and environmental sampling (WES) studies, reducing complexity and ensuring consistency across studies.
- Estimation of qPCR amplification efficiency: WES includes tools to estimate qPCR amplification efficiency, a critical parameter in validating qPCR-based detection methods.
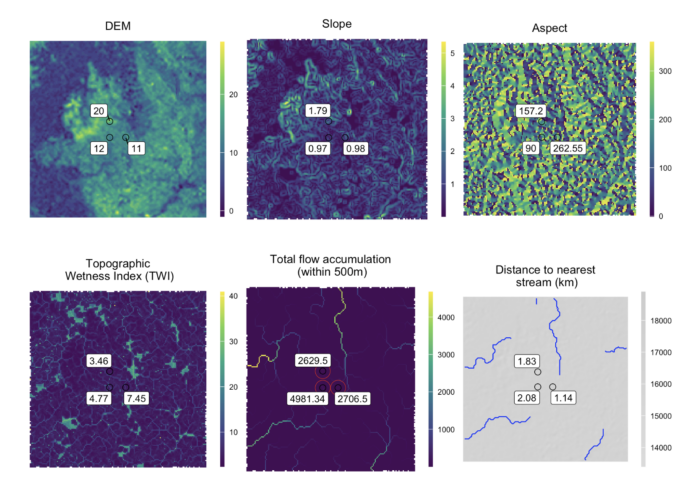
- Collation of open-source spatial data: The package assists with collating spatial data for sampling sites, integrating environmental and geographical information to enhance the analysis of environmental pathogen distribution.
Our approach
Our approach with the WES R package is to provide a user-friendly yet powerful toolkit for researchers and public health professionals engaged in environmental sampling studies. By offering standardized methods for data collation and analysis, the package ensures consistency and reproducibility across studies. The package is specifically tailored to support epidemiological surveillance in low- and middle-income countries (LMICs), where data is often collected from complex and informal sewer networks. We emphasize flexibility in application, ensuring that the tools can be adapted to various contexts that follow the same data structures.